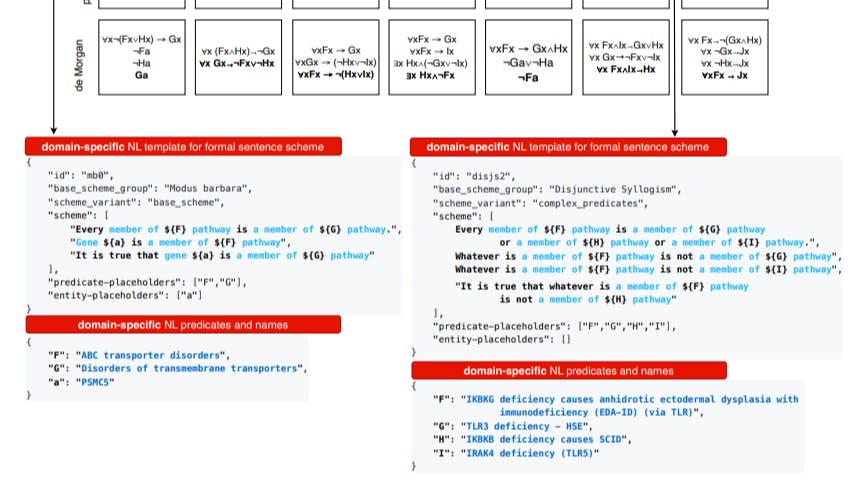
157 reads
Reactome Knowledgebase: Gene and Pathway Membership in Disease-Related Pathways
by
December 12th, 2024
Audio Presented by
The Large-ness of Large Language Models (LLMs) ushered in a technological revolution. We dissect the research.
Story's Credibility

About Author
The Large-ness of Large Language Models (LLMs) ushered in a technological revolution. We dissect the research.